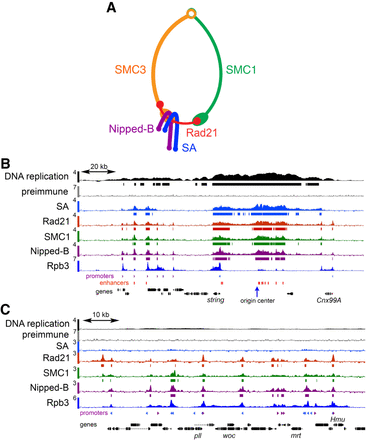
Cohesin and Nipped-B ChIP-seq in BG3 cells. (A) Cohesin subunit structure. (B) ChIP-seq near an early DNA replication origin at the string (stg, also known as cdc25) gene. ChIP-seq and DNA replication data are plotted as log2 enrichment. Bars below each track indicate enrichment in the 95th percentile over regions ≥300 bp. RpII33 (also known as Rpb3) RNA polymerase II subunit data are from a prior publication (Pherson et al. 2017). Locations of promoters (purple and blue, forward and reverse) and enhancers (red) are indicated below the tracks. (C) ChIP-seq in a region distant from an early replication origin containing the woc (without children) gene.











