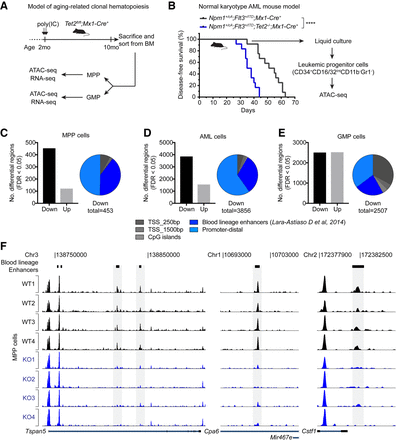
Differential analysis of chromatin accessibility reveals widespread changes in TF activity upon TET2 loss. (A) Diagram showing experimental setup to analyze native hematopoiesis in aged Tet2-deficient animals (see also Supplemental Fig. S3A). It has been demonstrated that native hematopoiesis is predominantly sustained by multipotent progenitors (MPPs) throughout the lifetime of the mouse (Sun et al. 2014; Busch et al. 2015). We therefore decided to perform genomic profiling on wild-type and Tet2-deficient MPPs as well as downstream myeloid-lineage progenitors (Granulocyte-Monocyte Progenitors [GMPs]). (B) Diagram showing experimental setup to analyze AML cells with and without deletion of Tet2. Genetically engineered mouse models (Lee et al. 2007; Quivoron et al. 2011; Vassiliou et al. 2011) carrying the indicated alleles were crossed and monitored for disease development. The Npm1+/cA;Flt3+/ITD genotype is sufficient to induce lethal AML (Mupo et al. 2013); however, ablation of Tet2 accelerates the onset of disease. Leaky Cre activity induces AML formation in the absence of pIpC-mediated induction of Mx1-Cre recombinase (Velasco-Hernandez et al. 2016). ATAC-seq analysis was performed on in vitro-grown leukemic progenitor cells isolated by FACS (see also Supplemental Fig. S3B). (****) P < 0.0001 (Log-rank [Mantel-Cox] test) in Kaplan-Meier plot. (C–E) Histogram showing the number of regions with significantly altered chromatin accessibility (FDR < 0.05) identified by diffBind analysis of wild-type and Tet2 knockout MPP cells (n = 4) (C), AML cells (n = 3) (D), and GMP cells (n = 4) (E). Pie charts indicate distribution of regions with decreased chromatin accessibility (down peaks) in Tet2 knockout cells with respect to promoters, CpG islands, and blood lineage enhancer regions (Lara-Astiaso et al. 2014). (F) Tracks of ATAC-seq signal in MPP cells from individual wild-type and Tet2 knockout animals (n = 4) at three representative genomic loci. Peaks that overlap a blood lineage enhancer with significantly decreased ATAC-seq reads in Tet2 knockout animals are highlighted in gray.











