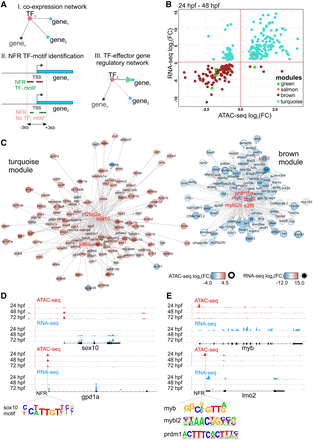
Dynamic regulatory networks of differentiating CMs. (A) Strategy used to establish gene-chromatin regulatory network. (B) Changes (log2FC) of gene expression compared to those in chromatin accessibility of cardiac module genes during heart development. Only significant (FDR < 0.05) genes are shown. (C) Regulatory networks of heart development. Arrows indicate the direction of interaction. Colors and the intensity of the circle edges indicate changes of chromatin accessibility, whereas those inside the circle show expression changes. Only significant (adjusted P-value ≤ 0.05) genes are shown. Hub TFs are indicated in red font. (D) Visualization of ATAC-seq and RNA-seq read coverage of selected genomic regions related to the turquoise module. (E) Visualization of ATAC-seq and RNA-seq read coverage of selected genomic regions related to the brown module. Time points, NFRs, and TF binding motifs within NFRs are indicated.











