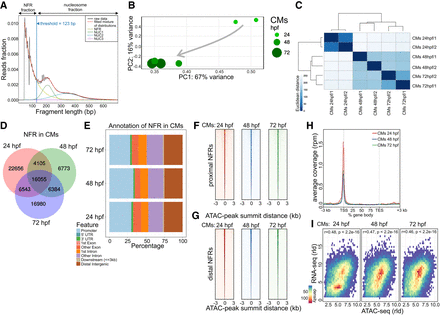
Cross-talk between transcriptome and chromatin accessibility profile across stages of cardiac development. (A) ATAC-seq read distribution and characterization of NFR fractions. (B) PCA of NFR chromatin accessibility during heart development. (C) Euclidian distances between chromatin accessibility within NFR. (D) Comparison of NFR presence and overlap across stages of heart development. (E) Genomic annotation of CM NFR consensus at different stages of heart development. (F) CM NFR consensus coverage heatmap of TSS proximal (±3 kb of TSS) regions centered on ATAC-seq peak summits. (G) CM NFR consensus coverage heatmap of TSS distal (more than ±3 kb of TSS) regions centered on ATAC-seq peak summits. (H) Metaplot of ATAC-seq read density over the gene bodies of the 1000 genes most highly expressed in CMs at each developmental stage. (TES) transcription end site. (I) Spearman's correlation of normalized log (rld) RNA-seq gene expression and ATAC-seq chromatin accessibility in corresponding NFR regions (±3 kb of TSS).











