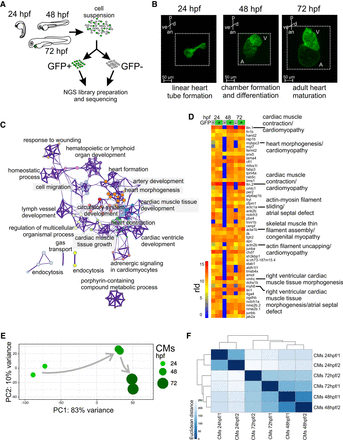
CM transcriptome landscape during heart development. (A) Schematics of experimental design. (B) Light sheet fluorescence microscope (LSFM) images of GFP-labeled CMs of developing zebrafish heart: (p) posterior; (an) anterior; (v) ventral; (d) dorsal. The dotted line indicates exact area of the LSFM image. (C) Network of 20 top-score GO clusters enriched in genes commonly up-regulated in GFP+ across heart development. Size nodes refer to the number of genes contributing to the same GO and nodes that share the same cluster ID are close to each other, adjusted P-value ≤ 0.05. (D) Heatmap of top 50 highly expressed genes between 24 and 72 hpf based on normalized expression value (regularized log [rld]). (E) Graphical representation of PCA of CM RNA-seq data. (F) Heatmap and clustering of RNA-seq sample-to-sample Euclidean distances.











