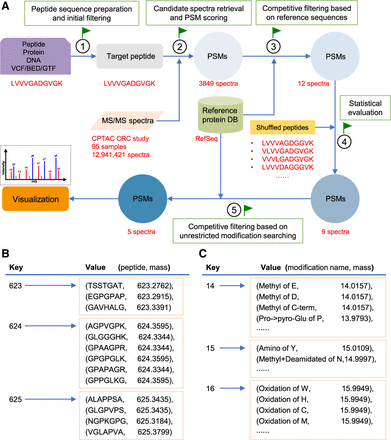
PepQuery workflow. (A) The PepQuery workflow involves five major steps: (1) target peptide sequence preparation and initial filtering; (2) candidate spectra retrieval and PSM scoring; (3) competitive filtering based on reference sequences; (4) statistical evaluation; and (5) competitive filtering based on unrestricted post-translational modification searching. The red text illustrates a real example in which a variant peptide LVVVGADGVGK is used to query the CPTAC colorectal cancer (CRC) data set with 95 samples and 12,941,421 spectra, using RefSeq as the reference protein database. Whereas existing methods require pairwise analysis between all 12,941,421 spectra and all RefSeq-derived peptide sequences plus the variant peptide sequence, PepQuery focuses only on the spectra that are relevant to the novel peptide, which reduces computational time and also allows more comprehensive analysis of these spectra. Illustration of peptide (B) and modification (C) indexing methods.











