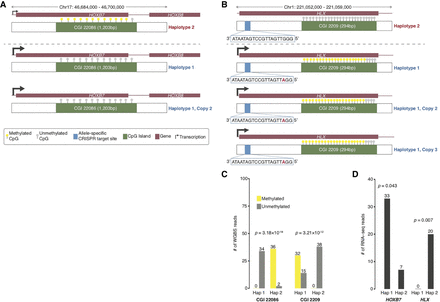
Genomic structural contexts provide insights into regulatory complexity. (A) Chr 17: 46,687,000–46,700,000 locus (triploid in K562) containing HOXB7 and HOXB8 and CpG Island (CGI) 22086 (1203 bp) where phased Haplotype 1 has two copies and Haplotype 2 has one copy. Allele-specific expression of HOXB7 from Haplotype 1. CpGs in CGI 22086 are unmethylated in Haplotype 1 and methylated in Haplotype 2. (B) Chr 1: 221,052,000–221,059,000 locus (tetraploid in K562) containing HLX and CGI 2209 (294 bp) where phased Haplotype 1 has three copies and Haplotype 2 has one copy. Allele-specific expression of HLX from Haplotype 1. CpGs in CGI 2209 are unmethylated in Haplotype 2 and highly methylated in Haplotype 1. Allele-specific CRISPR targeting site 797 bp inside the 5′ end of the HLX for both Haplotypes. (C) Number of methylated and unmethylated phased WGBS reads for Haplotypes 1 and 2 in CGI 22086 and CGI 2209 in which both CGIs exhibit allele-specific DNA methylation. (D) Number of RNA-seq reads for Haplotypes 1 and 2 of HLX and HOXB7 in which both genes exhibit allele-specific RNA expression.











