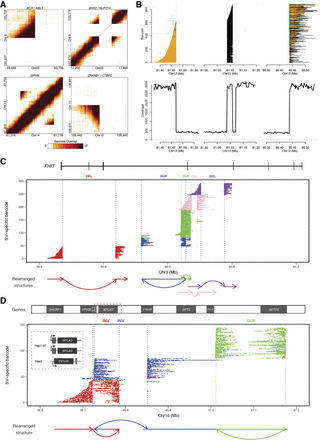
K562 SVs including large complex rearrangements resolved using linked-read sequencing. (A) Heat maps of overlapping barcodes for SVs in K562 resolved from linked-read sequencing using Long Ranger (Zheng et al. 2016; Marks et al. 2018). BCR/ABL1 translocation between Chromosomes 9 and 22. XKR3/NUP214 translocation between Chromosomes 9 and 22. Duplication within GPHN on Chromosome 14. Deletion that partially overlaps ZRANB1 and CTB2 on Chromosome 10. (B) Large complex rearrangement occurring on Chromosome 13 with informative reads from only one haplotype (region with loss of heterozygosity). Each line depicts a fragment inferred from linked reads based on clustering of identical barcodes (y-axis) using GROC-SVs (Spies et al. 2017). Abrupt endings (vertical dashed lines) of fragments indicate locations of breakpoints of this complex rearrangement. Fragments are phased locally with respect to surrounding SNVs (colored orange for same haplotype and black when no informative SNVs are found nearby). Gray lines indicate portions of fragments that do not support the current breakpoint. Fragments end abruptly at 81.47 Mb, indicating a breakpoint, picking up again at 81.09 Mb and continuing to 81.11 Mb where they end abruptly, then picking up again at 90.44 Mb. Coverage from 81.12 to 81.20 Mb are from reads with different sets of linked-read barcodes and thus are not part of this fragment set. (C,D) Complex rearrangements involving multiple haplotype-resolved SVs. Using gemtools (Greer et al. 2017), each SV is identified from linked reads grouped by identical barcodes (i.e., SV-specific barcodes, y-axis) indicative of single HMW DNA molecules (depicted by each row) that span the breakpoints. SVs are represented in different colors. The x-axis shows the hg19 genomic coordinate. Dotted lines represent individual breakpoints with schematic diagram of the rearranged structures drawn below the plot. (C) Multiple SVs within FHIT on 3p14.2. Deletion (DEL; red) (59.74–60.08 Mb) results in the loss of multiple exons. Two overlapping duplications (DUP; blue and green) in cis orientation (same allele of FHIT) indicated by the presence of HMW molecules spanning both DUPs. Two adjacent DELs (pink and purple) in trans (different alleles of FHIT), indicated by the absence of shared SV-specific barcodes for the HWM molecules spanning each DEL. SV haplotypes analyzed using SV-specific barcodes (not enough informative SNVs due to LOH). (D) Complex, intrachromosomal rearrangement spanning approximately 0.5 Mb on 16q11.2 and 16q12.1 that involve two overlapping inversions, 63 kb (red) and 125 kb (blue), and a 163-kb tandem duplication (green). This rearrangement resides on the nonduplicated haplotype of this triploid region. ORC6 is located entirely within the 63-kb inversion on 16q11.2 and is “deleted” by the left breakpoint of the 125-kb inversion, which also inverts MYLK3. C16orf8 on the same haplotype is also partially “deleted” by the 125-kb inversion (blue); NETO2 is duplicated by the 163-kb tandem duplication (green). (Inset) MYLK3 and ORC6 show allele-specific expression (Supplemental Table S12). MYLK3 is only expressed from this rearranged allele (Haplotype 2); ORC6 is expressed from the non-rearranged “diploid” allele (Haplotype 1).











