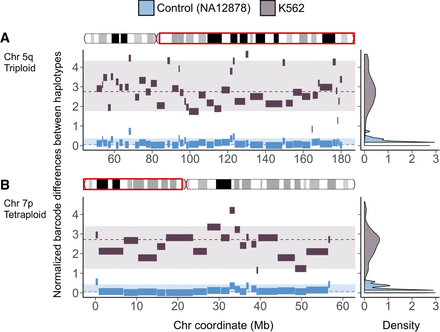
Mega-haplotypes of entire K562 chromosome arms: (x-axis) chromosome coordinate (Mb); (y-axis) difference in unique linked-read barcode counts between major and minor haplotypes, normalized for SNV density. Haplotype blocks from of normal control sample (NA12878) in blue and from K562 in dark gray. Density plots on the right reflect the distribution of the differences in haplotype-specific barcode counts for the control sample (blue) and K562 (dark gray). These density distributions are used for testing of significant difference (P < 0.001) using a one-sided t-test. Significant difference in haplotype-specific barcode counts indicates aneuploidy and haplotype imbalance. Haplotype blocks (with 100 or more phased SNVs) generated from Long Ranger (Supplemental Data Set S2) for the major and minor haplotypes were then “stitched” to mega-haplotypes encompassing the entire chromosome arm: (A) 5q (triploid); (B) 7p (tetraploid).











