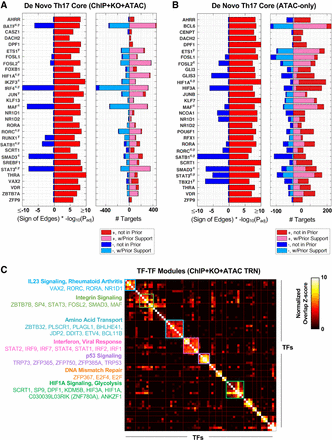
Analysis of the Th17 TRNs expands the “core” Th17 TRNs and predicts multivariate regulation of Th17 gene pathways. De novo Th17 core TFs in the ChIP + KO + ATAC TRN (A) and ATAC-only TRN (B). The Top 30 most significant “core” TFs are displayed. Significance was based on enrichment of a TF's (1) positive gene targets in up-regulated Th17 genes or (2) negative targets in down-regulated Th17 genes. (Left) Significance and direction of regulation; (right) number, sign, and prior support of TF target edges. Superscripts “c” and “y” indicate TF Th17 association from Ciofani et al. (2012) and Yosef et al. (2013), respectively. (C) Top 15 TF–TF modules for ChIP + KO + ATAC TRN. TFs were clustered into modules based on shared positive target genes between TFs (Methods). Gene-set enrichment was used to annotate clusters, and TF members are listed.











