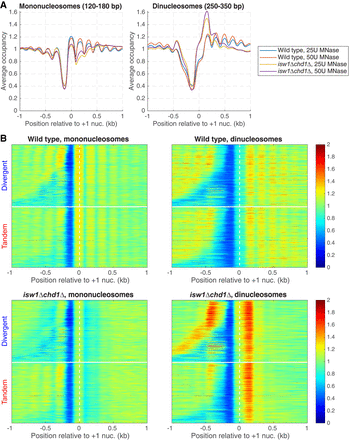
Closely packed dinucleosomes at the 5′ ends of genes in isw1Δ chd1Δ cells. MNase-seq data for wild-type and isw1Δ chd1Δ cells obtained without gel-purification of mononucleosomal DNA (two levels of digestion: 25 and 50 units MNase). (A) Nucleosome occupancy aggregate plots for mononucleosomes (120–180 bp) and dinucleosomes (250–350 bp) for all genes, aligned on the +1 nucleosome. Normalized to the genomic average (set at 1). (B) Heat maps for the data in A, separated into divergent and tandem gene pairs, sorted by intergenic distance and aligned on the +1 nucleosome of the downstream gene. The white horizontal line separates divergent from tandem gene pairs.











