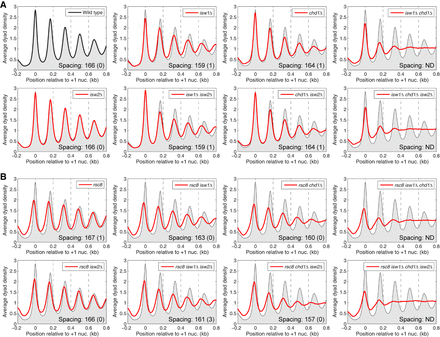
Combinatorial effects of RSC, ISW1, CHD1, and ISW2 remodelers on the chromatin organization of yeast genes. Average nucleosome phasing on all yeast genes relative to the dyad of the +1 nucleosome in wild-type cells. (A) Phasing in wild type, isw1Δ, chd1Δ, and isw2Δ mutants (from Ocampo et al. 2016). (B) Phasing in rsc8 mutants. For comparison, the phasing profile for wild-type cells is shown in all plots (gray shading). Inset values: average spacing determined by regression analysis of the first five peaks, with standard deviation (two biological replicate experiments); (ND) not determined because phasing is poor. (Note: Nucleosome spacing refers to the average distance between the centers of neighboring nucleosomes. An equivalent way of estimating the space between neighboring nucleosomes, or nucleosome repeat length, is by analyzing the periodicity of the bands observed after gel electrophoresis of MNase-digested chromatin.) The sequencing depths of all samples were normalized to one read per bp. See Supplemental Figure S1 for alignment on the TSS.











