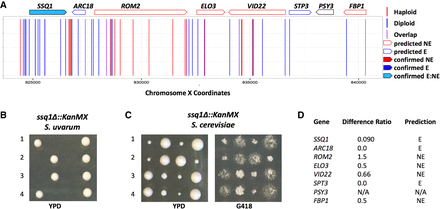
Validation of the S. uvarum–specific essential gene SSQ1. (A) Mapped chromosomal insertion positions are plotted across Chromosome X. Haploid inserts are indicated in red; diploid inserts, in blue; and overlapping inserts, in purple. Genes indicated across the top are outlined according to predicted dispensability and filled in if confirmed. Light blue filling indicates a gene that is essential in S. uvarum and nonessential in S. cerevisiae (confirmed E_NE). (B) Tetrad analysis of a heterozygous ssq1Δ::KanMX strain displaying inviable segregants containing the ssq1Δ allele in S. uvarum. (C) Tetrad analysis of a heterozygous ssq1Δ::KanMX strain in S. cerevisiae containing viable segregants plated on YPD and G418. (D) Table indicating the insertion ratio (number of haploid inserts by the number of diploid inserts) per gene. The final column lists the predicted classification: (NE) nonessential; (E) essential; (N/A) no data.











