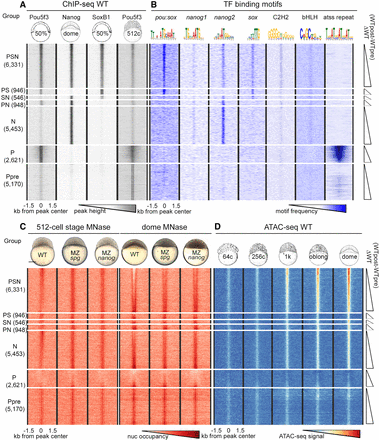
Nucleosomes cover all TFBSs pre-ZGA and are depleted from PSN triple-bound regions post-ZGA. Heat maps of six groups of genomic regions defined by combinatorial binding of Pou5f3, Nanog, and SoxB1 post-ZGA, as indicated at the left, and Ppre group bound by Pou5f3 only pre-ZGA. Three-kilobase genomic regions were aligned at the ChIP-seq peak centers. Within each group, the data were sorted by ascending difference between wild-type (WT) post-ZGA and pre-ZGA nucleosome occupancy (ΔWT post-pre). (A) TF binding (Pou5f3 50% epiboly, Nanog and SoxB1 dome stage, Pou5f3 512-cell stage). (B) Occurrence of TF binding motifs. (C) Nucleosome occupancy in the embryos of indicated genotypes. (D) Accessible chromatin signals in pre-ZGA (64c, 256c; c indicates cell stage), ZGA (1K), and post-ZGA stages (oblong, dome). (C) Scale bar, 200 µm.











