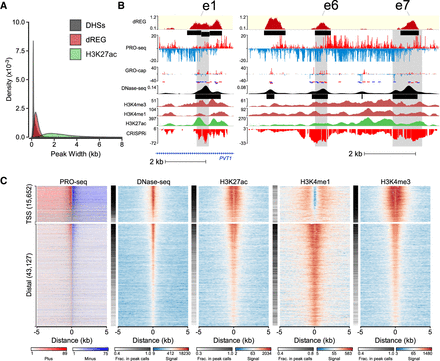
dREG calls are often concordant with other molecular assays. (A) Histogram shows the size distribution of dREG TIRs, H3K27ac ChIP-seq peaks, or DHSs. (B) WashU Epigenome Browser visualization of dREG signal, PRO-seq data, GRO-cap, DNase-seq, H3K4me3, H3K4me1, H3K27ac ChIP-seq, and CRISPR interference score (CRISPRi) at three enhancers (e1, e6, and e7) that affect transcription of MYC in K562 cells based on CRISPR interference (CRISPRi). (C) Heat maps show the log-signal intensity of PRO-seq, DNase-seq, or ChIP-seq for H3K27ac, H3K4me1, and H3K4me3. The fraction of sites intersecting ENCODE peak calls is shown in the white-black color map beside each plot. Color scales for signal and the fraction in peak calls are shown below the plot. Each row represents TIRs found overlapping an annotated transcription start site (n = 15,652) or >5 kb to a start site (n = 43,127).











