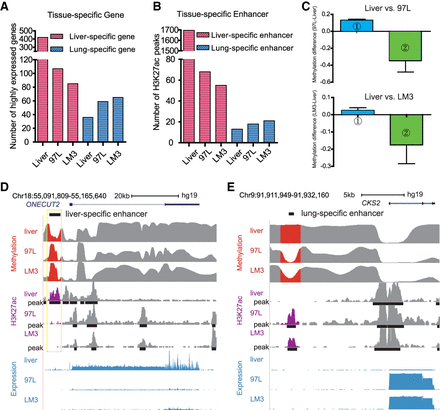
Aberrant DNA methylation pattern is correlated with enhancer alteration and cell identity. (A) 97L and LM3 lose the identity of liver by losing the liver-specific gene expression while they gain lung cell identity by expressing the lung-specific gene. The y-axis shows the number of highly expressed genes (FPKM ≥5). There are more lung-specific genes in LM3 cells compared with 97L cells. (B) Tissue-specific enhancers altered accordingly in 97L and LM3 with preferential metastasis to lung. For liver-specific enhancers, the number of overlapped H3K27ac peaks sequentially decrease in Liver, 97L, and LM3, which is related to loss of liver cell identity, whereas lung-specific enhancer is increased, especially in LM3 cells, which may promote expression of lung-specific genes, further helping LM3 cells metastasize to lung. (C) Loss or gain of tissue-specific enhancer is correlated with DNA methylation. Column 1 (blue bar) shows liver tissue-specific enhancers lost in hepatoma cells are accompanied by increased DNA methylation, and Column 2 (green bar) shows lung tissue-specific enhancers that are gained in hepatoma cells might be induced by largely decreased DNA methylation. The y-axis represents normalized and comparable methylation difference. Liver versus 97L is displayed in the upper panel, whereas the Liver versus LM3 is in the lower panel. (D) Tissue-specific enhancer gain or loss with aberrant DNA methylation is correlated with tissue-specific gene expression. Liver-specific gene ONECUT2 is down-regulated in 97L and LM3 with the loss of liver-specific enhancer where DNA methylation is increased. (E) Lung-related gene CKS2 is up-regulated in 97L and LM3 cells with gain of lung-specific enhancer where DNA methylation is decreased.











