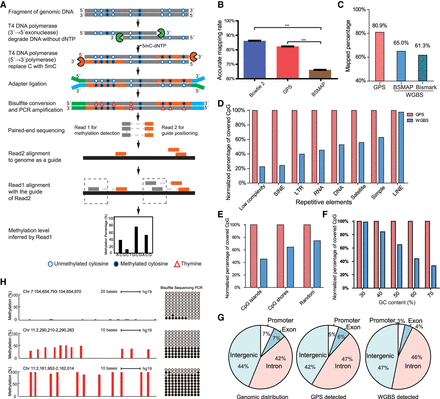
Guide Positioning Sequencing (GPS) detects genome-wide DNA methylation accurately with high coverage rate. (A) Schematic of GPS workflow for DNA methylation detection. The gray line represents original DNA sequence, and the orange line represents DNA treated by T4 DNA polymerase, which replaces cytosine with 5′-methylcytosine at 3′ end of DNA fragment. The solid circle (●) represents methylated cytosine, and the open circle (○) represents unmethylated cytosine, whereas the triangle (Δ) represents thymine. Blue and green short lines represent the NGS linker. Read1 represents the bisulfite-converted 5′ end of fragments, whereas Read2 represents the 3′ end of fragments, which is the same as the genome sequence due to 5′-methylcytosine replacement. (B) The accurate alignment rate of Bowtie 2 and GPS is obviously higher than that in BSMAP based on simulated data: (***) P < 0.001, one-tailed paired t-test. (C) Alignment efficiency of GPS compared with WGBS. DNA methylation of 97L cells was detected in parallel by GPS and WGBS, and mapped percentage of paired reads in GPS is 80.9%, 15%–20% higher than two popular WGBS alignment tools, BSMAP and Bismark. (D) Performance of GPS in detecting DNA methylation of repetitive elements. GPS had advantage over WGBS in detecting CpG sites of repetitive elements, especially in Low complexity, SINE, and LTR. The y-axis indicates the ratio (%) of WGBS/GPS-covered CpG sites. (E) Performance of GPS in CpG islands and CpG shores compared with WGBS. WGBS only detected ∼43% CpGs of those by GPS in CpG islands under similar sequencing depth for methylation detection. (F) Performance of GPS in different GC content. (G) Distributions of original genomic, GPS-detected, and WGBS-detected CpG sites. Genomic distribution shows total CpG sites in the genome. GPS detected that distribution of CpG sites is more similar to the original genomic CpG distribution compared with WGBS. (H) GPS-detected DNA methylation verified by Sanger sequencing from bisulfite-treated DNA. The red bar represents a single cytosine site, and the height of the red bar represents the methylation level. Each column of circles represents a single cytosine corresponding to the red bar. The solid circle (●) represents methylated cytosine, and the open circle (○) represents unmethylated cytosine.











