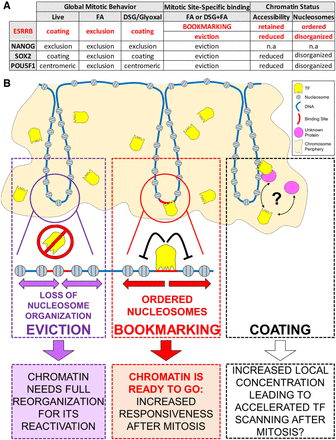
Model summarizing distinct behaviors of pluripotency TFs in mitotic cells and their relationships to nucleosome organization and post-mitotic gene regulation. (A) Summary table of the behavior of ESRRB, NANOG, SOX2, and POU5F1 in mitotic mouse ES cells. (B) Many TFs show global localization on the chromosomes in mitosis, such as ESRRB and SOX2. This localization is likely driven by sequence-independent interactions with DNA or other components of the chromatin or of the mitotic chromosomes and might serve a function in increasing the local concentration of TFs in proximity of their targets, in turn facilitating binding in G1. In contrast, during division, only few TFs remain dynamically bound to a subset of the sites they occupy in interphase, as exemplified by ESRRB. At bookmarked sites, the continued activity of these TFs maintains an ordered chromatin configuration, possibly limiting the extent of chromatin remodeling required to re-establish functional regulatory architectures in the following cell cycle. At sites losing TF binding, nucleosome positioning is disorganized, and increased occupancy by nucleosomes is detected at binding motifs. Although these sites do not become fully inaccessible, profound chromatin rearrangements are expected to be needed in early G1 to reinstate proper function.











