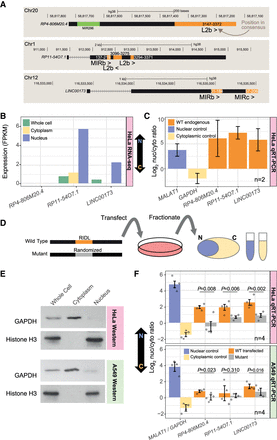
Disruption of RIDLs results in lncRNA relocalization from nucleus to cytoplasm. (A) Structures of candidate RIDL-lncRNAs. Orange indicates RIDL positions. For each RIDL, numbers indicate the position within the TE consensus, and its orientation with respect to the lncRNA is indicated by arrows. (>) same strand; (<) opposite strand. (B) Expression of the three lncRNA candidates as inferred from HeLa RNA-seq (Djebali et al. 2012). (C) Nuclear/cytoplasmic localization of endogenous candidate lncRNA copies in wild-type HeLa cells as measured by qRT-PCR. (D) Experimental design. (E) The purity of HeLa and A549 subcellular fractions was assessed by western blotting against specific markers. GAPDH/Histone H3 proteins are used as cytoplasmic/nuclear markers, respectively. (F) Nuclear/cytoplasmic localization of transfected candidate lncRNAs in HeLa (top) and A549 (bottom). GAPDH/MALAT1 are used as cytoplasmic/nuclear controls, respectively. (N) Number of biological replicates (values from all replicates are plotted; each replicate is represented by a different dot shape). Error bars, SEM. P-values for paired t-test (one tail) are shown.











