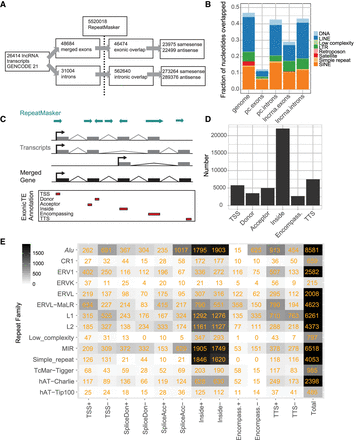
An exonic TE annotation with the GENCODE v21 lncRNA catalog. (A) Statistics for the exonic TE annotation process using GENCODE v21 lncRNAs. (B) The fraction of nucleotides overlapped by TEs for lncRNA exons and introns, protein-coding introns and exons (pc), and the whole genome. (C) Overview of the annotation process. The exons of all transcripts within a lncRNA gene annotation are merged. Merged exons are intersected with the RepeatMasker TE annotation. Intersecting TEs are classified into one of six categories (bottom) according to the gene structure with which they intersect and to the relative strand of the TE with respect to the gene: (TSS) overlapping the transcription start site; (donor) splice donor site; (acceptor) splice acceptor site; (inside) the TE boundaries both lie within the exon; (encompassing) the exon boundaries both lie within the TE; and (TTS) the transcription termination site. (D) Summary of classification breakdown for exonic TE annotation. (E) Classification of TE classes in exonic TE annotation. Numbers indicate instances of each type. (+ or −) Relative strand of the TE with respect to lncRNA transcript.











