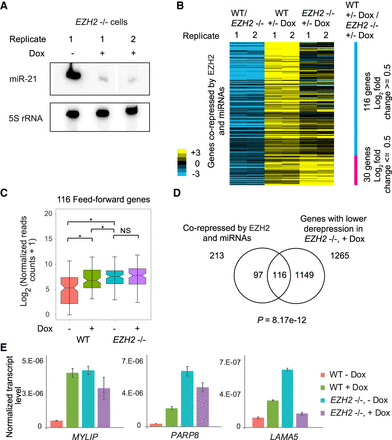
miRNAs repress PRC2 target genes through a feed-forward regulatory network with PRC2. (A) Depletion of miR-21 in response to VP55 induction in EZH2−/− cells. (B) Heat map showing derepression of gene expression in response to miRNA loss (+/− Dox) in WT (WT, +/− Dox), and EZH2−/− cells (EZH2−/−, +/− Dox) for genes corepressed by EZH2 and miRNAs. Only 146 of the 213 corepressed genes that had an average of 0.5 log2 fold difference in derepression between WT and EZH2−/− cells are shown. Genes are ordered in decreasing order of WT, +/− Dox/EZH2−/−, +/− Dox values. (C) Box plot comparing expression of feed-forward regulated genes in miRNA-depleted (+ Dox) and nondepleted (− Dox) WT and EZH2−/− cells. (*) P < 0.05; (NS) not significant. (D) Overlap between genes corepressed by EZH2 and miRNAs, and genes showing lower derepression upon miRNA depletion in EZH2−/− cells compared to WT cells. Significance of overlap was calculated using the hypergeometric test. (E) qRT-PCR showing transcript levels in miRNA-depleted WT and EZH2−/− cells, plotted as in Figure 1E.











