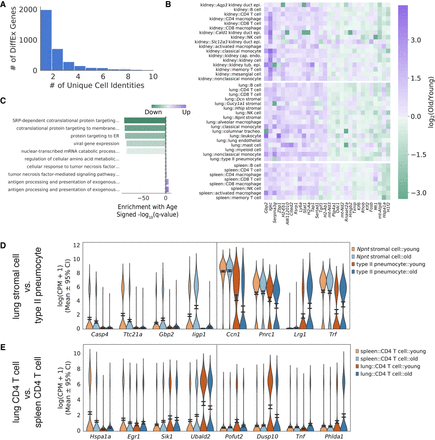
Differential expression analysis identifies common age-related changes across cell identities and tissue environments. (A) Distribution of the number of genes differentially expressed in at least k cell identities. We counted identical cell identities in different tissues (e.g., lung B cells, spleen B cells) as only one unique cell identity. We selected genes differentially expressed in k > 5 unique cell identities as common differentially expressed genes. (B) Heatmap of top 15 genes significantly changed in each direction across more than five cell identities. Fold changes between old and young cells are presented for each cell type in each tissue. Although no gene is universally changed across cell identities, each gene is changed across multiple tissues and developmental lineages. (C) Top five enriched Gene Ontology terms for genes that are down-regulated (negative values) and up-regulated (positive values) with aging across more than five cell identities. Dotted gray lines near 0 represent the α = 0.05 significance threshold. Antigen processing and metabolic pathways appear to be up-regulated, whereas protein translation and translocation pathways appear to be down-regulated with aging. (D) Violin plot of genes that are uniquely changed between two cell states in the lung (Wilcoxon rank-sum test, Q < 0.05): Npnt stromal cells and type II pneumocytes. Each gene presented is significantly up-regulated or down-regulated in one cell identity and does not change in the same direction (log2 [old/young] <0.1) in the other cell identity. Confidence intervals were computed by bootstrapping. For each cell state, we show the top two specific down-regulated genes and up-regulated genes. (E) Violin plot of genes that are uniquely changed between CD4 T cells isolated from the spleen and the lung. Genes were selected as in D.











