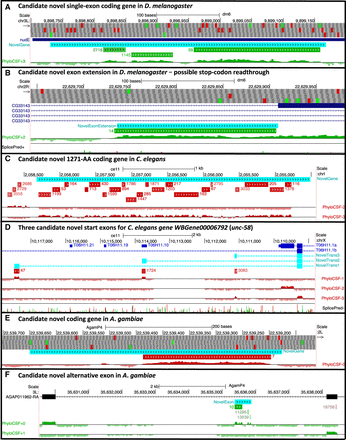
Potential novel CDSs in other species. Browser images show proposed novel CDSs (cyan) suggested by PCCRs (green/red for ± strand; rank next to region), smoothed PhyloCSF browser tracks, splice site predictions where useful (green donor, red acceptor, height indicating prediction strength), and ATG (green) and stop (red) codons. Supplemental Figure S6 has color-coded alignments for each example. (A) A cluster of three PCCRs in the 5′ UTR of D. melanogaster nudE suggest there is a single-exon novel protein-coding gene or an additional nudE cistron with ORF at positions 9898731–9899168. Although there is no PhyloCSF signal in the first 28 codons, the high frame conservation despite several indels provides ample evidence of purifying selection for protein-coding function. (B) A PCCR just 5′ of an exon of D. melanogaster transcript F of CG33143 suggests that there is a novel coding transcript including an exon 173 nt longer than the annotated exon. This exon includes an in-frame TAG stop codon, suggesting translational stop codon readthrough. We have previously estimated that ∼6% of D. melanogaster genes undergo stop codon readthrough (Jungreis et al. 2016). The stop codon is perfectly conserved and is followed immediately by a cytosine residue, both of which are known correlates of readthrough. (C) A large cluster of PCCRs on the “−” strand of C. elegans Chromosome I suggests there is a 1271-amino-acid single-exon gene with ORF at positions 2054512–2058327. There is no alignment for a few codons on each end of the PhyloCSF signal, so to construct the putative ORF, we have extended the region 5′ to the nearest ATG and 3′ to the nearest stop codon. (D) Three PCCRs within an intron of C. elegans gene WBGene00006792 (unc-58) shown on the “−” strand of Chromosome X suggest alternative start exons for that gene. The coding region of each of these putative exons begins with a perfectly conserved ATG and ends at a perfectly conserved GT having high splice-prediction score. All three end with a 1-nt partial codon, which allows them to splice to the next exon of transcript T06H11.1b while preserving the reading frame. (E) A PCCR in A. gambiae suggests that 22539177–22539650 on the “−” strand of Chromosome 2L is protein coding, forming either a novel gene or the first coding exon of the previously incompletely annotated gene AGAP005849. Subsequent curation confirmed the latter. Frame conservation provides strong evidence of coding function in the early portion of the putative transcript where the PhyloCSF signal is weak. (F) A cluster of three PCCRs in an intron of A. gambiae gene AGAP011962 suggests an additional coding exon at positions 35635374–35635874 of Chromosome 3L, confirmed through subsequent curation to be part of a previously missed alternative transcript.











