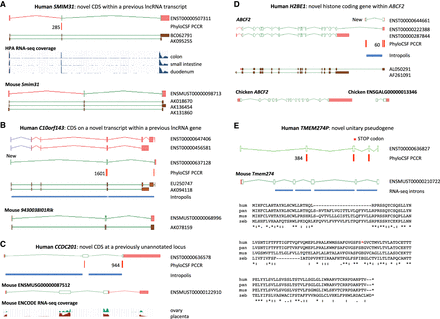
Novel protein-coding loci. Browser images show CDSs (open green rectangles), UTRs (pink), supporting PCCRs (red), top rank (black), cDNA evidence (brown), and RNA-seq–supported introns (blue rectangles). Additional transcript models omitted for clarity. Multispecies protein alignments showing conservation of complete ORFs are in Supplemental Figure S4. (A) Novel coding gene SMIM31, previously a cDNA-supported GENCODE lincRNA, was changed to protein coding without a change of transcript structure owing to a 71-aa CDS (ENST00000507311) conserved to coelacanth. The protein-coding cDNA-supported ortholog was added to mouse GENCODE (Smim31). PhyloCSF does not detect coding potential in the second coding exon, but multispecies protein alignment and preponderance of 3-mer indels provide evidence this exon is coding. Human Protein Atlas (HPA) RNA-seq and human and mouse FANTOM5 CAGE data show high transcription in gastrointestinal tissues. (B) Novel coding gene C10orf143 was previously a GENCODE lncRNA (LINC00959), with two cDNA-derived models (ENST00000647406 and ENST00000456581). Discovery of the 108-aa CDS required adding a transcript model (ENST00000637128), supported by Intropolis short-read data. The original lncRNA transcripts have been reannotated as nonsense-mediated decay targets (purple ORFs), based on a premature stop codon in a cassette exon. The orthologous cDNA-supported mouse locus had previously been recognized as protein coding (9430038I01Rik). The gene has a broad expression profile in both species. (C) CCDC201 is a novel human gene with a 187-aa CDS conserved to birds, previously missed owing to lack of spliced cDNA or EST evidence. The ancestral stop codon has been lost in rodents, adding a 30-aa extension in novel mouse protein-coding gene ENSMUSG00000087512. Introns are supported by Intropolis short-read RNA-seq, limited to female reproductive tissues and certain developmental cells. Mouse ENCODE RNA-seq supports placenta and ovary expression only, and the mouse locus (in the guise of a ncRNA) had previously been identified as a target for the germ cell–specific transcription factor Figla (Joshi et al. 2007). (D) H2BE1 is a novel histone HB2 family member protein-coding gene with a 122-aa CDS (model ENST00000644661), whose first exon was identified in this study. Intropolis supports the transcript structure, with expression limited to oocytes and embryonic cells (e.g., SRR499827). Human FANTOM5 CAGE data lacks experiments from developmental stages, which may explain the absence of TSS evidence. Overlapping model ENST00000222388 had previously been annotated as an alternative transcript of ABCF2 (ancestral CDS represented by model ENST00000287844) based on cDNA AL050291, with putative translation in the shared exon following the coding frame of ABCF2. PhyloCSF indicates that the 122-aa CDS is translated in a different frame, so the translation of ENST00000222388 is potentially spurious. Although the 122-aa CDS is conserved to birds, the locus has apparently been lost in rodents. There is no evidence for transcriptional connectivity between the orthologous Ensembl chicken models ABCF2 and ENSGALG00000013346 (bottom). ENST00000222388 has been reclassified as a “readthrough” transcript, and Intropolis data indicate that such readthrough between human ABCF2 and H2BE1 is rare. (E) TMEM274P is a novel human unitary pseudogene, orthologous to novel mouse protein-coding gene Tmem274. CDS alignments to RefSeq models such as scallop LOC110448246 and trichoplax XP_002113670.1 suggest this gene may predate vertebrate evolution, although orthology is presumptive owing to lack of synteny beyond coelacanth. The gene has at best weak expression data in all species examined, but all but one of the mouse splice junctions is supported by minimal ENCODE RNA-seq data from pooled sources, and all splice sites display mammalian conservation. An alignment of human (hum) to chimp (pan), with outgroups mouse (mus) and zebrafish (zeb), shows that human has a premature stop codon that is not a known SNP in the fourth exon of the ancestral CDS (red asterisk in diagram and alignment) and has also lost the second coding exon (large gap in human sequence); both events are unique to human. The zebrafish sequence in the alignment is from XP_017212190, and the chimp translation is from the genome sequence.











