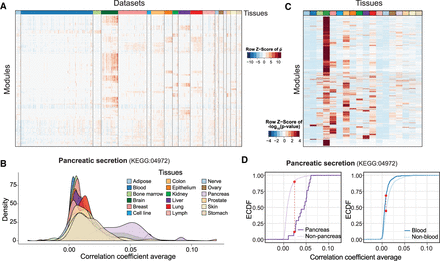
Predicting tissue specificity of modules. (A) Heat map showing the correlation coefficient averages of genes 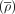 in modules from expression data of a subset of human data sets. Data sets from different tissues are arranged and colored
(top bar). Modules are clustered in rows using hierarchical clustering.
in modules from expression data of a subset of human data sets. Data sets from different tissues are arranged and colored
(top bar). Modules are clustered in rows using hierarchical clustering. 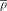 values for each module are centered and scaled. (B) Coexpressions among genes of pancreatic secretion module across tissues in human. The average correlation coefficients across
the genes in the pancreatic secretion module in human data sets are used to illustrate the coexpressions of this module across
tissues. Genes in the pancreatic secretion module have higher coexpression in data sets from the pancreas compared to those
from other tissues. (C) Heat map showing the tissue specificity of modules inferred from the correlation coefficient of respective tissues against
the other tissues. Modules are clustered in rows using hierarchical clustering. The −log10(P-values) obtained from the K–S test are centered and scaled for each module. (D) The tissue-specificity of pancreatic secretion in pancreas (left) and blood (right) is illustrated by the empirical cumulative distribution function (ECDF). The red dotted lines indicate the K–S statistic,
which is based on the maximum distance between the two curves. Curves shifting toward the right indicate that data sets from the respective tissue have a higher correlation coefficient and, therefore, greater specificity
for this tissue. In this case, the steeply rising part of the ECDF, also shown as the peak of the density of the correlations
in B, is shifted toward higher correlations.
values for each module are centered and scaled. (B) Coexpressions among genes of pancreatic secretion module across tissues in human. The average correlation coefficients across
the genes in the pancreatic secretion module in human data sets are used to illustrate the coexpressions of this module across
tissues. Genes in the pancreatic secretion module have higher coexpression in data sets from the pancreas compared to those
from other tissues. (C) Heat map showing the tissue specificity of modules inferred from the correlation coefficient of respective tissues against
the other tissues. Modules are clustered in rows using hierarchical clustering. The −log10(P-values) obtained from the K–S test are centered and scaled for each module. (D) The tissue-specificity of pancreatic secretion in pancreas (left) and blood (right) is illustrated by the empirical cumulative distribution function (ECDF). The red dotted lines indicate the K–S statistic,
which is based on the maximum distance between the two curves. Curves shifting toward the right indicate that data sets from the respective tissue have a higher correlation coefficient and, therefore, greater specificity
for this tissue. In this case, the steeply rising part of the ECDF, also shown as the peak of the density of the correlations
in B, is shifted toward higher correlations.











