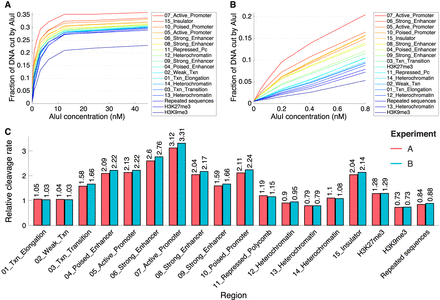
Heterochromatin and euchromatin have very similar DNA accessibilities in mouse hepatocytes. (A) AluI digestion kinetics for sites in the 15 epigenetic chromatin states in mouse hepatocytes defined by Bogu et al. (2016), for sites in repeated sequences (defined by RepeatMasker) and for sites marked by H3K9me3 (constitutive heterochromatin) or H3K27me3 (facultative heterochromatin) (data from Nicetto et al. 2019). (B) Initial rates of AluI digestion. (C) Quantitative comparison of initial digestion rates for the 15 epigenetic states defined by Bogu et al. (2016), repeated sequences, and regions marked by H3K9me3 or H3K27me3. Data for biological replicate experiments A and B are shown. See Supplemental Material for details of the analysis.











