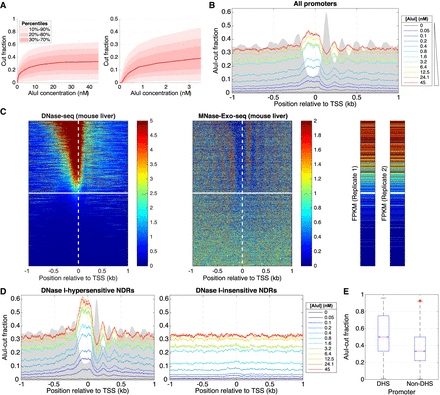
Inactive gene promoters are accessible to AluI in some mouse liver cells. (A) AluI digestion of mouse hepatocyte nuclei (12 digestion points): (left) all data; (right) initial digestion. The data range shows that 80% of AluI sites (with at least five reads) are cut in 5%–60% of cells. (B) Average AluI accessibility plotted as a function of distance from the TSS on all approximately 24,000 mouse genes. Gray area indicates MNase-Exo-seq data (nucleosome dyads) (Cole et al. 2016) on an arbitrary scale. (C) Heat map analysis of all approximately 24,000 mouse genes sorted according to the DNase I hypersensitivity of their promoters in mouse hepatocytes (data from ENCODE) and aligned on the TSS: (left) DNase I cut density; (middle) nucleosome dyad positions (Cole et al. 2016); (right) RNA-seq data (two biological replicates from ENCODE). The white line divides hypersensitive and insensitive promoters (defined in Supplemental Fig. S5). (D) Average AluI accessibility plotted as a function of distance from the TSS for DNase I–hypersensitive and DNase I–insensitive promoters defined in C. (E) Distribution of AluI-cut fractions corresponding to all AluI sites located in promoters (region [TSS − 185 bp; TSS + 85 bp]), separated by DNase I hypersensitivity.











