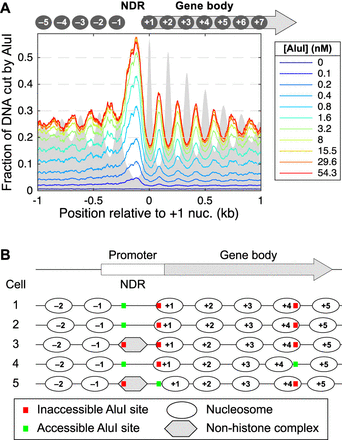
Genomic analysis of AluI accessibility reveals imperfect nucleosome phasing in yeast. (A) Mean accessibility as a function of distance from the center of the +1 nucleosome (defined by Chereji et al. 2018) on all approximately 5000 yeast genes. (B) Heterogeneous nucleosome positioning model to explain the AluI accessibility data. On a typical gene, nucleosomes are positioned slightly differently in each cell such that a particular AluI site is inside a nucleosome in one cell and in a linker in another cell. The cartoon shows the nucleosome positions on a gene in five different cells. An AluI site in the coding region is in the linker (accessible) in only one cell out of five (20% accessibility), whereas an AluI site in the promoter NDR is accessible in three out of five cells (60% accessibility). The observed average values are ∼25% in the coding region and ∼55% in the NDR (see A).











