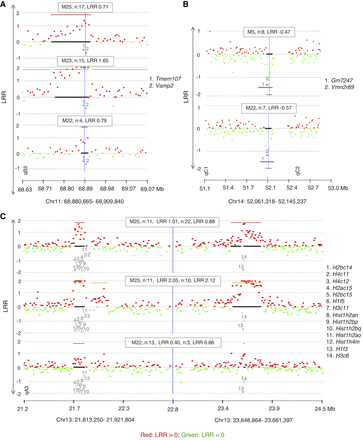
CGH array results showing recurrent CNAs in memory cells from PLNs of diabetic mice (n: number of affected probes, location of genes is indicated by numbers). (A) A recurrent copy gain in cells of three mice spanning several genes including Tmem107. (B) Recurrent copy loss spanning two undocumented genes (Gm7247 and Vmn2r89). (C) An independent recurrence of copy gain in three different mice that are spanning two Histone families’ loci. The formula to calculate the percentage of mosaicism of deletions is 100 ×(1–2LRR) × 2 (example: the percentage of mosaicism for −0.25 is 31.8% mosaicism and for −0.5 is 58.6%). DLRSpread values of samples are in Table 1 and Supplemental Fig. S2.











