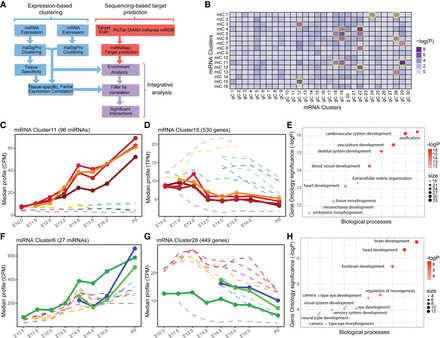
Identification of miRNA–mRNA cluster interactions. (A) Potential targets of each miRNA cluster were obtained by applying an ensemble approach. Interactions were called as significant if they had a negative tissue-specific partial correlation and were enriched beyond the Bonferroni-corrected P-value of 10−4. (B) Heatmap of miRNA cluster target enrichment calculated using χ2 statistics. The 18 interactions identified as enriched are boxed in orange and gold. The interactions boxed in gold have negative partial correlation and are identified as significant interactions. (C) miRNA cluster 11 corresponds to brain-specific miRNAs up-regulated during development. (D) mRNA cluster 18 genes are highly expressed in other organs such as limbs, cranioface, and heart. (E) Gene Ontology of miRNA cluster 11 targets in mRNA cluster 18 shows enrichments in developmentally important genes with roles outside the brain. (F) miRNA cluster 6 increases significantly during heart development. (G) mRNA cluster 28 genes are overexpressed in the brain. (H) Gene Ontology analysis of miRNA cluster 6 targets in mRNA cluster 28 revealed terms such as brain, head, and forebrain development.











