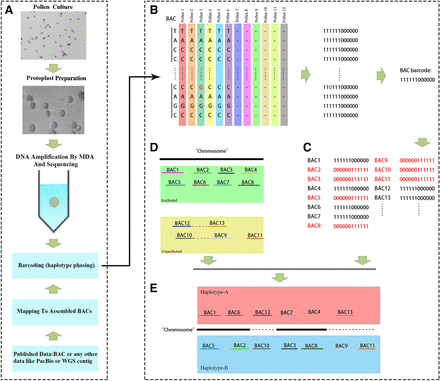
Schematic of haploid phasing using a barcoding approach. (A) Pollen is a haploid gamete. Here, we combined a new approach for pollen protoplast isolation with a single-cell DNA amplification technique and then used a barcoding bioinformatics strategy to incorporate haploid-specific sequence data from pollen cells, ultimately enabling the efficient and accurate phasing to assemble a haplotype genome as well as detecting meiotic recombination events. (B) Establishing the relationship between 12 pollen cells and BAC sequences using 12-bit binary codes in which “1” indicates identical and “0” indicates “not identical” or “absent.” Each base was labeled with a 12-bit binary code. Finally, each BAC was labeled with a specific 12-bit binary code. (C) BACs from different haplotypes received different 12-bit binary codes. (D) The chromosomal location of each BAC on the reference genome chromosomes was determined by aligning the BACs to the anchored reference chromosomes. (E) BACs were classified into haplotypes based on the chromosomal location and the 12-bit binary code.











