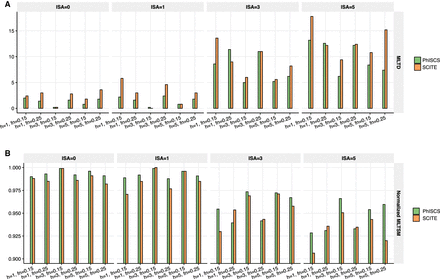
Comparison of PhISCS with SCITE on simulated data with multiple bulk samples and larger number of subclones and mutations. In each case, 10 distinct trees of tumor evolution were generated with 15 subclones and 100 mutations (SNVs). The number of cells was set to 100, while the depth of coverage for bulk data was set to 5000×. A compares the two tools with respect to the MLTD dissimilarity measure, while B compares them with respect to its dual MLTSM similarity measure (normalized values of MLTSM are shown in the figure). On the x-axes, h, fn, and ISA, respectively, denote the number of bulk samples, false negative rate of single-cell data, and the number of simulated mutations for which ISA is violated.











