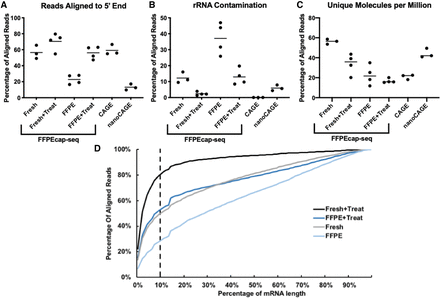
Enzymatic pretreatment improves library quality. Quality metrics, including percentage of reads mapping to the 10% most 5′ end of transcripts (A), percentage of reads mapping to rRNAs (B), and the percentage of unique reads from a random sampling of 1 million reads (C), are comparable to CAGE and nanoCAGE libraries and show improvement following enzymatic treatments for libraries made with either freshly derived or FFPE-derived RNA. Lines in graph represent means. (D) The cumulative percentage of reads within a certain percentage of mRNA length, starting at the 5′ end, is shown.











