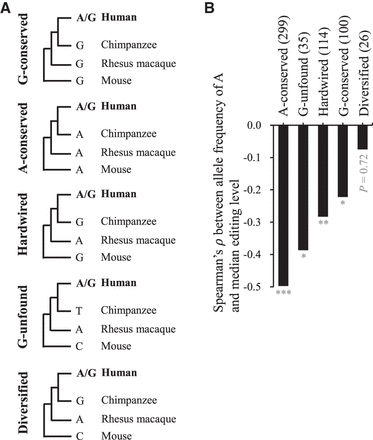
Phylogenetic analysis of nonsynonymous A-to-G editing sites. (A) Definition of five types of human nonsynonymous editing sites (A-conserved, G-unfound, hardwired, G-conserved, and diversified) based on their evolutionary variations among human, chimpanzee, rhesus macaque, and mouse. (B) Correlations between median editing levels and allele frequency of A within the LCL population for the five groups of editing sites. The number of nonsynonymous editing sites examined in each group is provided in parentheses. P-values were determined using a two-tailed Wilcoxon rank-sum test. (*) P-value < 0.05, (**) P-value < 0.01, (***) P-value < 0.001.











