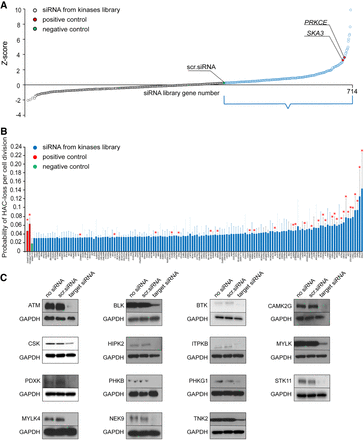
Mitotic stability of HAC/dGFP in human HT1080 cells treated with siRNA library against protein kinases. (A) Scatter plot showing a distribution of siRNAs against protein kinases based on Z-score. The genes marked in blue and positioned between a negative control (scr. siRNA in green) and the highest score are suitable for the next step of the analysis. SKA3 and PRKCE were used as positive controls (red). (B) Mitotic stability of HAC/dGFP in human HT1080 cells treated with siRNA library against protein kinases. siRNAs against SKA3 and PRKCE were used as positive controls, and a scrambled siRNA (scr. siRNA) as a negative control. Among 714 genes analyzed, the strongest effect on HAC loss was shown by siRNAs against ITPKB, IRAK1, MYLK, TNK2, STK38, BLK, MAPK7, FRK, TRIO, STK11, CRIM1, CSK, PDXK, PHKG1, KSR2, CAMK2G, PHKB, CSNK1G2, MYLK4, TPD52L3, RBKS, TTBK1, PNCK, PINK1, BTK, HIPK2, BUB1, ATM, BUB1B, TAOK1, and NEK9. Red asterisks indicate statistical significance (P < 0.05; t-test) when compared to a negative control. (C) Western blot analysis confirming silencing efficiency of ATM, BLK, BTK, CAMK2G, CSK, HIPK2, ITPKB, MYLK, PDXK, PHKB, PHKG1, STK11, MYLK4, NEK9, and TNK2 proteins (Supplemental Table S8) after siRNA-mediated knockdown.











