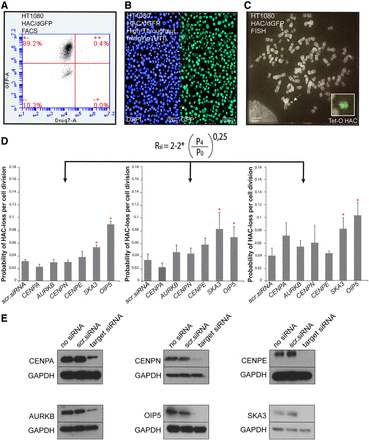
Validation of the HAC/dGFP-HTI assay. Measurement of the proportion of GFP-positive and GFP-negative cells in HT1080 human cells carrying HAC/dGFP treated with a set of siRNAs designed to knockdown known genes essential for chromosomal transmission using flow cytometry (FACS) (A), HTI (B), and fluorescence in situ hybridization (FISH) (C). (D) The formula used to determine the rate of HAC loss per generation after siRNA treatment (Methods). (A–C) The rate of HAC/dGFP loss after treatment with a set of siRNAs against CENPA, AURKB, CENPN, SKA3, and OIP5 as measured by FACS (A), HTI (B), and FISH (C). The error bars indicate standard deviation. Knockdown of SKA3 and OIP5 shows the highest effect on HAC/dGFP loss. Red asterisks indicate siRNA treatment that results in statistically significant difference (P < 0.05; t-test) when compared to a negative control (scr. siRNA or nontargeting siRNA). (E) Western blot analysis confirming silencing efficiency of CENPA, CENPN, CENPE, AURKB, OIP5, and SKA3 proteins (Supplemental Table S8) after siRNA-mediated knockdown of the genes.











