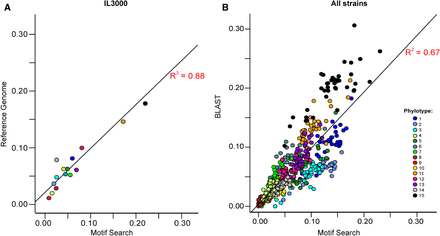
Performance of the protein motif-based variant antigen profile (VAP). (A) Correlation of motif-based and manually curated phylotype frequencies in the T. congolense IL3000 reference genome sequence. Pearson's product moment correlation statistics: R2= 0.88, t(13) = 9.7321, P < 0.001. (B) Correlation of motif-based and manually curated phylotype frequencies in 41 T. congolense strains. Manual VAPs were estimated by counting the top matches from BLASTx (Altschul et al. 1990). Pearson's product moment correlation: R2= 0.64, t(566) = 34.39, P < 0.001. Phylotypes are color-coded according to the key.











