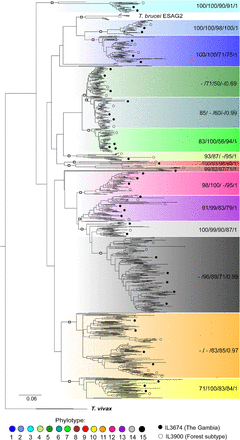
Maximum likelihood (ML) phylogeny of T. congolense VSGs. The phylogeny was estimated from full-length VSG protein sequences of IL3000 (Kenya), IL3674 (The Gambia), and IL3900 (Forest subtype, Burkina Faso) with RAxML (Stamatakis 2014) using a ML method with a WAG+Γ model and 100 bootstrap replicates. The 15 phylotypes identified in IL3000 are color-coded according to the key. Positions of example sequences from IL3674 and IL3900 are indicated according to the key. Labels for the internal nodes of each phylotype (marked by the open squares) are shown on the right. These labels indicate the bootstrap percentages for ML from the complete tree (RAxML), as well as ML (PhyML) (Guindon et al. 2010), ML (MEGA7) (Kumar et al. 2016), neighbor joining (NJ) (Felsenstein 1989), and posterior probabilities (BI) (Huelsenbeck and Ronquist 2001) estimated from a pruned tree containing 147 sequences. Tree is rooted with two T. vivax VSG sequences (Fam23).











