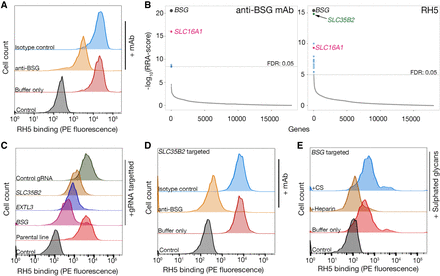
Identification of BSG and heparan sulfate as independent receptors for P. falciparum RH5 on HEK293 cells. (A) Biotinylated RH5 was clustered around a streptavidin–PE conjugate and binding to HEK293 cells was analyzed by flow cytometry. RH5 binding is only partially reduced by a blocking anti-BSG mAb relative to controls. (B) Rank-ordered genes identified from gRNA enrichment analysis required for cell surface display of an anti-BSG mAb (left) and RH5 binding (right). Significantly enriched genes with a FDR < 0.05 are colored (full screening results available in Supplemental Data S2); genes encoding the receptor (BSG) and chaperone (SLC16A1) were common to both screens, and a gene involved in GAG-biosynthesis (SLC35B2) was additionally required for RH5 binding. (C) Binding of RH5 to cells is reduced when transduced with lentiviruses encoding gRNAs targeting either the receptor (BSG) or enzymes required for HS synthesis (SLC35B2, EXTL3) relative to controls. Transduced polyclonal lines were used for this experiment. (D) RH5 binding to SLC35B2-targeted HEK293 cells could be completely prevented if preincubated with a blocking anti-BSG mAb but not an isotype-matched control. (E) RH5 binding to BSG-targeted HEK293 cells could be completely blocked if preincubated with 200 µg/mL heparin but not 200 µg/mL CS. A representative of three independent (A and C) or technical (D and E) replicate experiments is shown.











