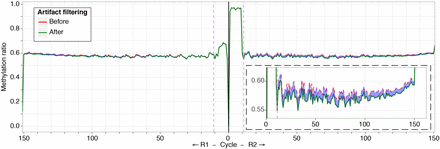
A subset of reads shows apparent cytosine methylation throughout their lengths, in CG and CH dinucleotide contexts. Our use of 5mC for end repair reveals the expected ∼10-bp increase in the C/T ratio (y-axis) in read 1 (right). However, prompted by our finding of unconverted cytosines in lambda DNA, we tested whether any of the reads had a pattern of unconverted cytosines in all dinucleotide contexts (the expected CG and the uncommon CH contexts). We found a subset of reads in which no cytosines were converted. To categorize these, we developed the filterFillIn2 software to identify four consecutive unconverted cytosines in a CHH context. The effect on DNA methylation values is shown in the C/T plots, the original values in red and following removal in green. The inset shows that ∼1%–2% of the DNA methylation value is accounted for by this artifact.











