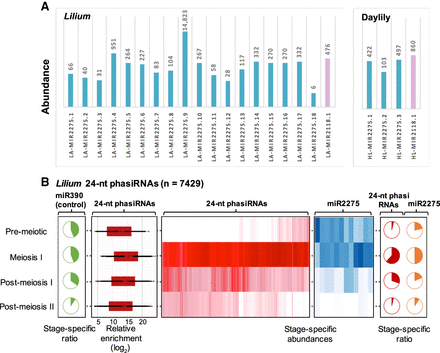
Reproductive PHAS triggers and 24-nt phasiRNAs in Lilium. (A) miR2118 (violet) and miR2275 (blue) family members identified in Lilium and daylily by comparing mature sRNA sequences to members in miRBase (version 21); matches to variants scoring ≤4 were considered as valid candidates. Variants include mismatches between mature sRNAs and miRNAs from miRBASE, plus 5′ or 3′ overhang in the alignment. Values above the bars represent their total abundance (TP30M) in anthers. (B) Heat maps depicting abundance of Lilium 24-nt phasiRNAs (red) and miR2275 triggers (blue) in developing anthers. Both heat maps are clustered on similarity of expression. Pie charts represent the proportion of stage-specific abundances for 24-nt phasiRNAs (red), miR2275 (orange), and miR390 (green), the trigger of tasiRNAs across different anther developmental stages that are included in this study. Box-whisker plot shows enrichment (log2) of Lilium 24-nt phasiRNA abundance from all PHAS loci in the meiotic anther compared to the vegetative sample (leaf).











