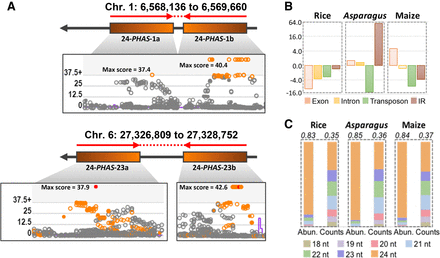
Many Asparagus 24-PHAS loci are derived from inverted repeats. (A) Genomic organization of two representative IR-type PHAS loci, overlapping with the 5′ and 3′ arm of inverted repeats. Phasing scores from sliding window analysis are presented as shown in our custom web viewer. Loci with score >25 were considered as phased loci. (B) Fold change representing enrichment or depletion of overlap of 24-PHAS loci from rice, Asparagus, and maize, with exons, introns, transposons, and inverted repeats, versus random chance. The complete data are in Supplemental Table S5. (C) Comparison of sRNA abundances and unique sRNA counts of sRNAs produced from 24-PHAS loci from rice, Asparagus, and maize. The values on top represent 2.5% trimmed mean of ratio of abundances or counts of 24-nt phasiRNAs from all 24-PHAS loci of corresponding species.











