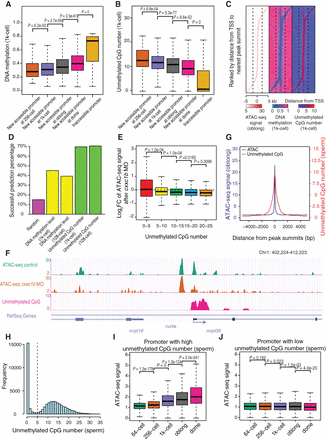
Unmethylated CpGs prime the emergence of accessible promoters. (A) The box plot shows the relationship between newly accessible promoters and DNA methylation levels (1k-cell stage). Earlier accessible promoters have lower DNA methylation levels. P-values were calculated by Wilcoxon test. (B) The box plot shows the relationship between newly accessible promoters and the largest unmethylated CpG numbers among 200-bp bins (1k-cell stage). Earlier accessible promoters have higher numbers of unmethylated CpGs. P-values were calculated by Wilcoxon test. (C) The heat map shows the relationship between the locations of the ATAC-seq peak, DNA methylation level (1k-cell stage), and unmethylated CpG number (1k-cell stage) at the promoters. The accessible promoters in C–E are newly emerged ones in the 1k-cell or oblong stages. (D) The bar plot shows the successful prediction percentage of the locations of the ATAC-seq peak summits at the promoters, based on the locally lowest DNA methylation level or the local highest number of unmethylated CpGs (128-cell and 1k-cell stages, respectively). Details of the prediction of the ATAC-seq peak summit locations were described in Supplemental Methods. (E) The box plot shows the log2-transformed fold change in the ATAC-seq signals at the 200-bp bins with largest numbers of unmethylated CpGs at each accessible promoter upon knockdown of cxxc1b. The accessible promoters were grouped based on the locally largest numbers of unmethylated CpGs. P-values were calculated by Wilcoxon test. (F) The genome browser view shows the ATAC-seq signals at the mrpl39 locus as a representative example of decreasing accessibility at the unmethylated CpG-enriched regions upon knockdown of cxxc1b. (G) The summit of the ATAC-seq signals overlaps with the highest number of unmethylated CpGs in the sperm at the promoters. The ATAC-seq peaks are newly emerged ones in 1k-cell or oblong stages. (H) All promoters are divided into two groups according to the locally largest numbers of unmethylated CpGs in sperm (cut-off of 5). (I) The box plot shows the ATAC-seq signal during ZGA at the promoters with a high locally largest number of unmethylated CpGs in sperm. P-values were calculated by Wilcoxon test. (J) The box plot shows the ATAC-seq signal during ZGA at the promoters with a low locally largest number of unmethylated CpGs in sperm. P-values were calculated by Wilcoxon test.











