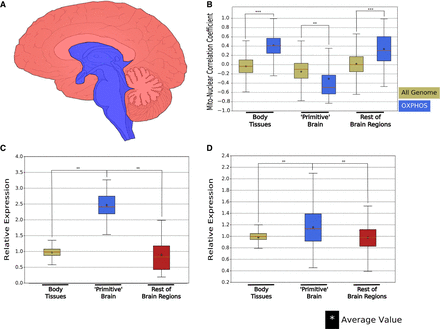
“Primitive” brain reveals a negative mito-nuclear expression pattern and an overall increase in OXPHOS gene expression levels. (A) A schematic illustration of human brain cross-section, roughly dividing the brain into associated with the “primitive” (blue) or other brain regions (red). (B) A box plot representing Spearman's correlation coefficients between either the entire genome (greenish gray) or nDNA-encoded OXPHOS genes with mtDNA genes (blue) in all nonbrain body sites, in an aggregate of all “primitive” brain regions (that each showed negative mito-nuclear OXPHOS genes correlations, separately) and in all other brain regions as well (see Supplemental Data Sets S1, S2). Red lines: median correlation coefficient; stars: average correlation coefficient. (***) P < 1 × 10−100. (C,D) Box plots representing the expression of mtDNA-encoded (C) and nDNA-encoded (D) OXPHOS genes relative to the median expression across all 48 tested tissues, in all nonbrain body sites (greenish-gray), in an aggregate of all “primitive” brain regions that showed (separately) negative mito-nuclear OXPHOS genes correlations (blue), and in all other brain tissues (red) (see Supplemental Data Sets S1, S2). Red lines: median correlation coefficient; stars: average correlation coefficient. (**) P < 1 × 10−10.











