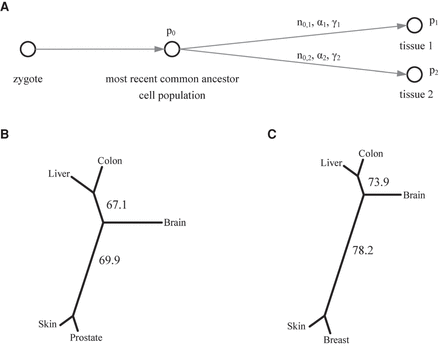
Measuring inter-tissue distance based on the AF similarity of shared mosaicism. (A) The AF difference of shared mosaicisms in two tissues could be traced back to their most recent common ancestral cell population, but not to an earlier stage when the mosaicisms were generated. (B,C) Clustering trees based on the pairwise distance matrix estimated from the WGS data set for (B) males and (C) females. The bootstrapping values labeled on the internal branches show the bootstrap supporting percentage on the partition of that branch.











