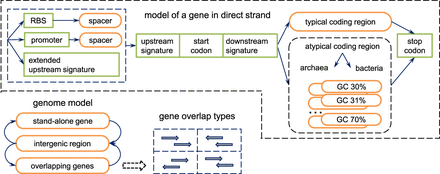
Principal state diagram of the generalized hidden Markov model (GHHM) of prokaryotic genomic sequence. States shown in the top panel were used to model a gene in the direct strand. Genes in the reverse strand were modeled by the identical set of states (with directions of transition reversed). The states modeling genes in direct and reverse strands were connected through the intergenic region state as well as the states of genes overlapping in opposite strands.











