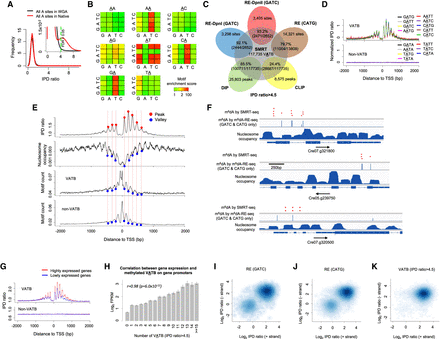
Characterization of a complete m6dA methylome of C. reinhardtii reveals novel biological insights. (A) FDR estimation by comparing the IPD ratio distribution of C. reinhardtii native (red) with WGA (black) samples. The inset provides an enlarged view. (B) A rigorous motif enrichment analysis reveals that VATB (V = A, C, or G and B = C, G, or T) is the m6dA motif of in C. reinhardtii. Each 4 × 4 heatmap corresponds to all 16 4-mer motifs, for which the second and third bases are fixed at the center/title (e.g., AA). The rows and columns in the heatmaps represent the first and last bases of 4-mer motifs. Each cell in the following 4 × 4 heatmaps shows the motif enrichment score based on the native DNA sample. (C) Putative m6dA sites called by SMRT-seq are highly consistent with those detected by independent techniques: m6dA-DIP-seq (DIP), m6dA-CLIP-exo-seq (CLIP), and m6dA-RE-seq (RE). (D) VATB, but not non-VATB (i.e., TATN/NATA), motifs have a periodic pattern of IPD ratio distribution around TSSs. Average IPD ratio (normalized by motif frequency) for each of the nine VATB motifs (top) and each of the seven non-VATB motifs (bottom) are plotted around TSSs. (E) Relationship across four different distributions (top to bottom panels): average IPD ratio of VATB sites, nucleosome positioning, and frequency of VATB and non-VATB motif sites. Peaks and valleys of the periodic patterns are indicated by red and blue dots, aligned across the four panels. (F) Illustrative examples showing m6dA sites near the TSSs of three genes. This figure is adapted from Fu et al. (2015), where we project m6dA sites detected by SMRT-seq (red dots; FDR < 0.05; randomly generated heights to ease visualization) on top of GATC and CATG sites detected by m6dA-RE-seq (blue bars; middle) and nucleosome occupancy (bottom). (G) m6dA events at VATB sites are associated with active gene expression. Average IPD ratios are compared between two groups of genes with high (FPKM > 1) and low (FPKM < 1) expression levels. (H) The correlation between the gene expression level in C. reinhardtii and methylated VATB on gene promoters. The x-axis represents the number of methylated VATB sites (IPD ratio > 4.5; FDR = 0.05) within [0, +2000 bp] of TSSs. The y-axis represents the mean log2 FPKM of genes. Error bars, SEs. (I–K) Single-molecule, strand-specific analysis of SMRT-seq data to examine full-, non-, or hemi-methylation status at m6dA sites. Three sets of m6dA sites are analyzed: m6dA in GATC sites (I) and CATG sites (J) based on m6dA-RE-seq (Fu et al. 2015) and (K) VATB sites with high aggregate IPD ratio (IPD ratio > 4.5; Methods) based on SMRT-seq. The x- and y-axes denote the single-molecule, strand-specific IPD ratio of each pair of reverse-complementary VATB sites at the two strands of each single molecule.











