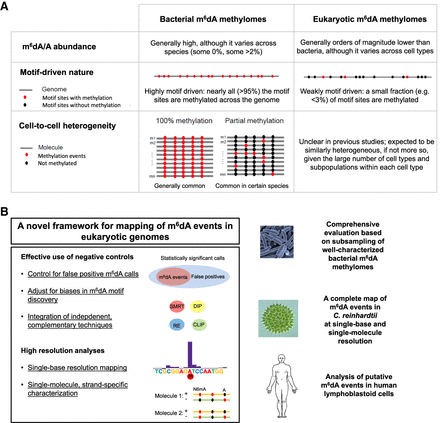
Differences between bacterial and eukaryotic m6dA methylomes and a novel approach for mapping m6dA events in eukaryotic organisms. (A) Comparison between bacterial and eukaryotic m6dA methylomes over three aspects. (B) A novel approach for mapping and characterizing m6dA events in eukaryotic genomes. The novel approach, including a set of methods as summarized on the left, is comprehensively evaluated using subsampled bacterial m6dA methylome data and applied to Chlamydomonas reinhardtii (green algae) and human lymphoblastoid cells (LCLs).











