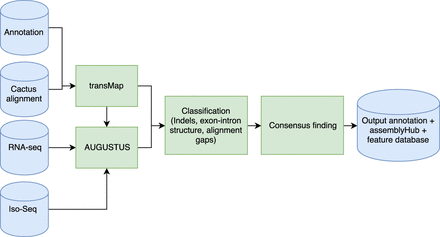
CAT pipeline schematic. The CAT pipeline takes as input a HAL alignment file, an existing annotation set, and aligned RNA-seq reads. CAT uses the Cactus alignment to project annotations to other genomes using transMap (Stanke et al. 2008). These transcript projections are then filtered and paralog resolved. Optionally, AUGUSTUS can be run in as many as four parameterizations. All transcripts are classified for extrinsic support and structure, and a “chooser” algorithm picks the best representative for each input transcript, incorporating ab initio transcripts when they provide novel supported information. The final consensus gene set, as well as associated feature tracks, are used to create an assembly hub ready to be loaded by the UCSC Genome Browser (for more detail, see Supplemental Fig. S1; Supplemental Methods).











