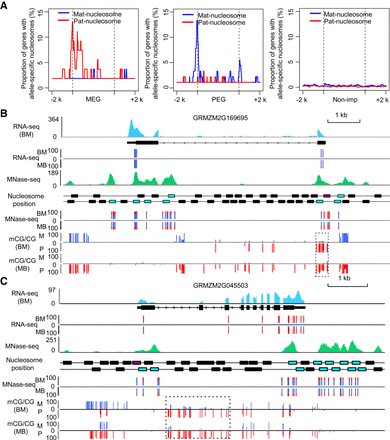
Relationship between imprinted genes and parent-of-origin-dependent nucleosomes. (A) The distribution of parent-of-origin nucleosomes at imprinted genes/nonimprinted genes and its up and down 2-kb regions. The blue line represents the maternal specifically positioned nucleosome (mat-nucleosome), and the red line represents the paternal specifically positioned nucleosome (pat-nucleosome). Nonimprinted genes were expressed in endosperm and did not deviate significantly from the 2:1 ratio of maternal allele to paternal allele in both reciprocal hybrids. (B,C) The investigated view of expression, DNA methylation, and nucleosome occupancy at GRMZM2G169695 (B) and GRMZM2G045503 (C). The expression levels of transcribed regions are shown in light blue. The normalized nucleosome levels of endosperm (BM) are plotted in green. The black, blue, pink, and purple rectangles indicate the position of endosperm nucleosomes, allelically analyzed nucleosomes, low-stringency allele-specific nucleosomes, and high-stringency allele-specific nucleosomes at genes. The percentages of allelic reads of RNA-seq and MNase-seq data for specific SNP sites are shown, with red lines for the paternal allele and blue lines for the maternal allele. The DNA methylation level for specific SNP sites are shown for both maternal and paternal alleles, with red lines for the paternal allele (P) and blue lines for the maternal allele (M). The dotted rectangles highlight the pDMRs identified in this region.











